My Digital Garden Setup
Table of Contents
Digital Garden is a collection of inter-linked notes on the web, that like a garden, is constantly growing and evolving. It is a way to learn in the open.
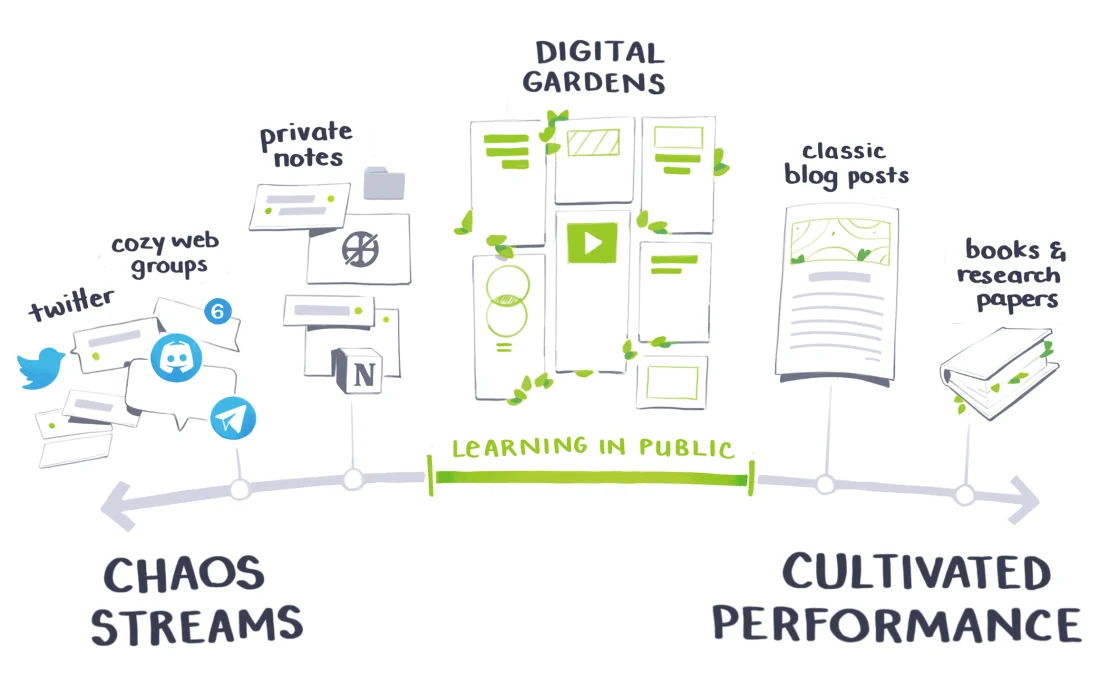
Figure 1: Learning in Public (maggieappleton.com)
I started writing digital notes from 2020. I didn't have a digital garden back then, just some notes in .org files in my computer. Then, one day in August 2023, I published it all in my website. I was inspired by seeing Jethro Kuan's braindump website.
So, why do I write, and why should you write?
- Writing is thinking: The process of writing, wording out things, giving a structure to it in a text file forces you to organize your thoughts and think clearly.
Writing is remembering: Or at least you can reabsorb the information more quickly next time.
But still, even if you don't read your notes, writing helps in understanding.
Learning in the open: You don't need to know everything, your writing doesn't need to be perfect. Write about the things your are learning and doing.
Make the thing you wish you had found when you were learning. Don't write for claps and likes, write for yourself from 3 months ago.
1. How is this site built
The site is made up of notes, each page is an individual .org file, that link among themselves and other websites on the internet. It is hosted on github pages and the domain is handled by Cloudflare. Here are the steps to go from .org files to a website:
- I use org roam to manage my notes in Emacs,
- Then org's built in org-publish mechanism exports those notes to html,
I have some scripts to build the sitemap, rss file and the graph shown in the index page [/].
I took the graph analysis code from Hugo Cisneros and modified his graph rendering code to add some interactive zoom, pan and physics .
- Another script goes through all the html files and adds CSS and JS inside the <head> tag, and does some cleanup.
- Finally, I give a look at the changes, commit them, and push to github pages.
To edit and view my notes in my phone, I use orgro. And to sync my notes I use the fabulous syncthing program.
You could take my setup and adapt it to your needs. For that, you'd need to install Emacs, get familiar with org roam. Running .lisp scripts will require installing SBCL compiler and quicklisp package manager. And you are all set to go.
2. How can you have your own site
First step is choosing a tool and writing notes. I'd suggest going with Org-mode in Emacs or with Obsidian. One benefit with Obsidian is that it is easier to get started with and also has good support for mobile devices. Org-mode on the other hand is more flexible and feature rich. Both of them provide ways to publish to html. Don't stress about which tools to use, the main point is to write notes.
I didn't mention Notion above because we want our notes to last for lifetime. And I believe the best way to ensures that is to keep them in plain text format, in your own device. Also, I suggest practising a good habit of doing backups regularly.
3. Appendix
3.1. Switching tools
It can seem like a big deal but don't stress about which tools to use, the main point is to write notes. If you decide later to switch tools, pandoc can convert between them and if necessary you could write a script that fixes any issues.
I didn't have to switch tools (like from Obsidian to Emacs), but within org files too I changed directory structure, link format and few other things. It is a journey and I figured out what works and doesn't work as I went along. One of the benefits of having it in text file is that you can bulk edit the files with simple scripts.
3.2. org-publish
The following code configures org-publish to take org files from my notes folder and output html files in my website folder:
(setq org-publish-project-alist `(("braindump" :base-directory "~/Documents/notes/" :base-extension "org" :publishing-directory "~/Development/Website/braindump/" :exclude "^notes.org\\|^tasks.org\\|^rss.org" :recursive nil :headline-levels 4 :auto-preamble t :html-prefer-user-labels t :auto-sitemap nil)))
Then M-x org-publish braindump exports all my notes to as html.
3.3. Scripts
The overall flow and scripts are:
- generate_sitemap.lisp generates the sitemap file
- Inside emacs
org-publishpublishes all the notes and sitemap to html file - generate_rss.lisp generates the rss file
generate_graph_data.py generates .json file with information required to show the graph of notes in the index page [/].
In the index page graph.js fetches that .json file and renders the graph.
- Finally, format-html.lisp goes through all the html files and adds CSS and JS inside the <head> tag, and does some cleanup.
Of course, I don't got through this step by step everytime. I have a shell script that runs all this.